Installing optimap¶
optimap is a Python library for optical mapping analysis that supports Windows, Mac OSX, and Linux. This guide will help you get started with installation and basic usage.
optimap requires Python 3.8 or later. To install Python we recommend installing the Anaconda distribution, which includes Python and many useful packages for scientific computing, or by installing Python directly.
Tip
optimap relies heavily on NumPy and Matplotlib. For an introduction to Python and these libraries, we recommend:
Installation using pip (macOS, Windows, Linux)¶
The easiest way to install optimap is using pip in the command line:
pip install optimap
To update optimap to the latest version run
pip install --upgrade optimap
Installing without Recommended Dependencies
pip install optimap will install optimap with all recommended dependencies including OpenCV (opencv-contrib-python) and PySide6, which may not be desired in some advanced use cases. If you need a custom version of OpenCV (e.g., with CUDA support for GPU-accelerated motion tracking) or a different Qt implementation use the following command to install optimap with minimal dependencies:
pip install --no-deps optimap
pip install -r https://raw.githubusercontent.com/cardiacvision/optimap/refs/heads/main/requirements-core.txt
or using pip-mark-installed:
pip install pip-mark-installed
pip-mark-installed opencv-contrib-python PySide6
pip install optimap
Installing from source¶
To install optimap from source, clone the GitHub repository and run pip install . in the root directory:
git clone https://github.com/cardiacvision/optimap.git
cd optimap
pip install .
See Contributing for more details.
Overview of optimap¶
optimap is organized into the following modules:
optimap - An open-source Python toolbox for processing optical mapping and fluorescence imaging data. |
|
Functions for loading, viewing, filtering, saving and exporting videos. |
|
Functions for loading, saving, and displaying images, and for creating masks. |
|
Functions for extracting time-series from videos. |
|
Motion estimation and compensation functions for optical mapping data. |
|
Activation map computation module. |
|
Functions for computing, filtering, and analyzing phase maps from videos, and for detecting phase singularities. |
|
General utility functions. |
The most important functions are also imported into the top-level namespace optimap. Click on the module name to see the functions and classes it contains.
See the tutorials listed below for a comprehensive introduction to optimap’s main features. For a complete reference of all available functions and classes, consult the 📚 API documentation.
- Tutorials
- Tutorial 0: Overview of optimap
- Tutorial 1: Basics
- Tutorial 2: Import / Export (I/O)
- Tutorial 3: Measurement of Fluorescent Signals and Waves
- Tutorial 4: Working with Image Masks
- Tutorial 5: Motion Compensation
- Tutorial 6: Activation Maps and Beat Detection
- Tutorial 7: Conduction Velocity
- Tutorial 8: Ratiometry
- Tutorial 9: Action Potential Duration
- Tutorial 10: Phase and Singularities during VF
- Tutorial 12: Plotting Optical Traces
- Tutorial 13: Smoothing
- Example Data Files
Using optimap¶
We highly recommend using Visual Studio Code for working with optimap. Visual Studio Code is a free and open-source editor with excellent support for Python and Jupyter notebooks.
To get started:
Download and install Visual Studio Code
Install the Python extension by Microsoft
Install the Jupyter extension by Microsoft
Code Cells in VS Code¶
VS Code allows you to create interactive code cells directly in Python files by typing # %%. This lets you run code segments individually, similar to Jupyter notebooks or MATLAB Sections:

For more details, see the VS Code documentation on Jupyter support. Similar code cell functionality is also available in other editors like PyCharm and Spyder.

Command Line Usage¶
If you prefer using the command line, you can:
Run scripts directly with
python3 your_script.pyfrom the script’s directoryStart an interactive Python session by running
python3and enter commands directly
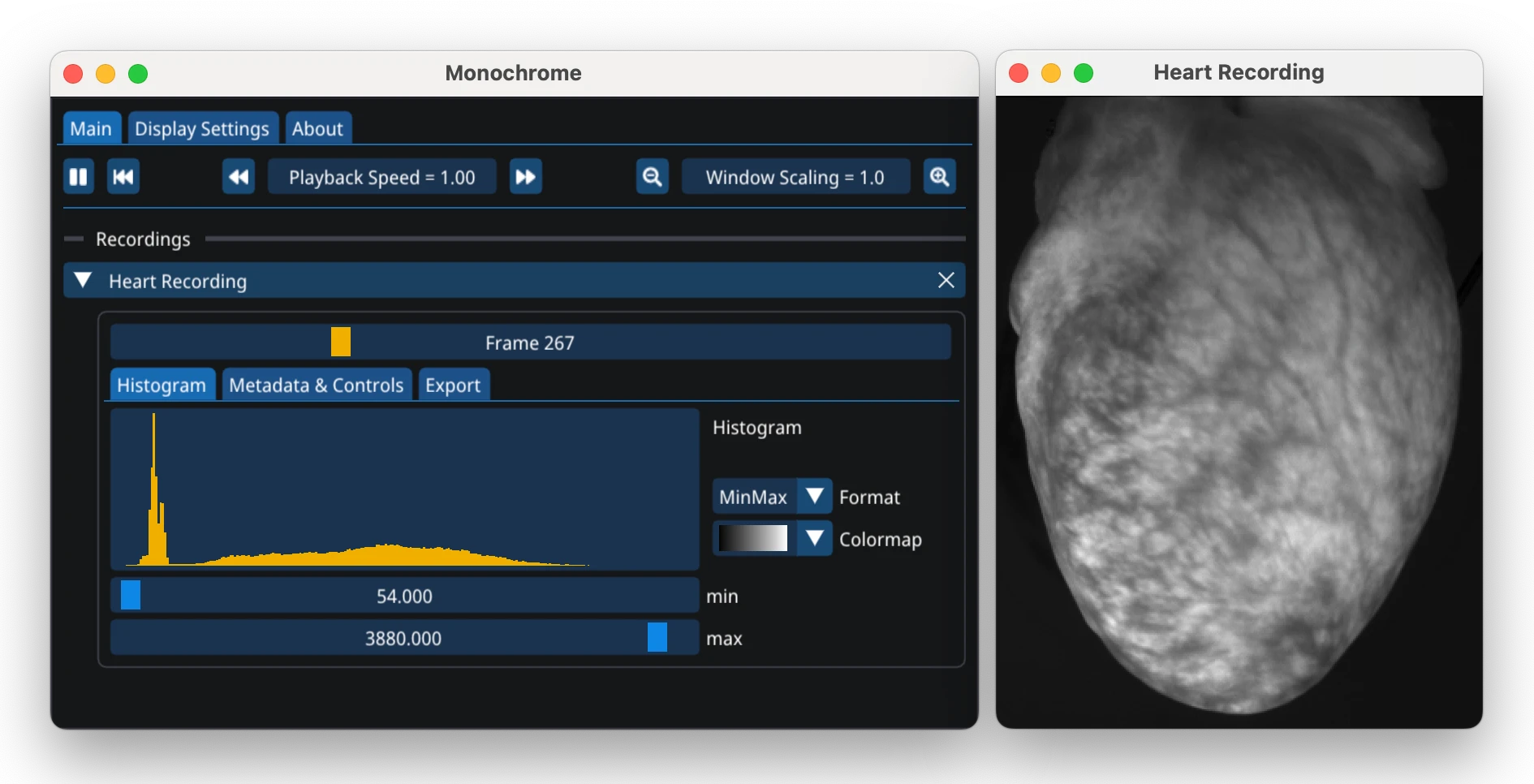
Monochrome Viewer¶
Monochrome is a companion tool for viewing high-dynamic range monochromatic videos, commonly used in optical mapping. While not required for optimap, it provides additional visualization capabilities. To view a video in Monochrome use the following Python code:
import monochrome as mc
mc.show(video)
See the Monochrome documentation for details.
Interactive Plots¶
Working with remote Jupyter notebooks
The interactive plotting functions in optimap may not work as expected when using Jupyter Notebook on a remote server (i.e., not on your local computer). This is because the interactive plots require a local display to work properly.
optimap uses Matplotlib for plotting. The following functions require an interactive Matplotlib backend:
|
Simple video player based on matplotlib's animate function. |
|
Video player for n side-by-side videos based on matplotlib's animate function. |
|
Video player for two videos side-by-side based on matplotlib's animate function. |
|
Play a video with an overlay. |
|
Interactive selection of positions on an image. |
|
Interactive selection/plotting of traces from a video. |
|
Compare traces of multiple videos. |
|
Simple video player with displacement vectors displayed as arrows. |
Simple video player with displacement vectors displayed as points. |
We strongly recommend using the Qt matplotlib backend for interactive plotting. optimap will attempt to automatically switch to the Qt backend for these functions and switch back to the inline backend afterwards (if applicable).
Note
optimap.utils.disable_interactive_backend_switching() can be used to disable the interactive backend switching if it causes problems. Please report any issues you encounter.
Alternatively, you can manually switch to the Qt backend by running the following code before calling any of the functions listed above:
import matplotlib.pyplot as plt
plt.switch_backend("QtAgg") # or "QtCairo"
How to cite¶
A publication about optimap is currently in preparation. In the meantime, please cite the following DOI and paper if you use optimap in your research: Lebert and Christoph[1], Lebert et al.[2].